
|
PostParser a tool to embrace myriads of DNA |
PostParser - an instrument to analyze data sets of DNA.
PostParser is a special biological software developed to work with nucleotide sequences. Most of todays molecular biology
experiments deal with DNA or RNA structures. Information of primary structure can be retrieved from high-throughput sequencing or classical Sanger method.
In the first case, hundred thousand sequences (reads, contigs) can be obtained, but subsequent clarification of the biological significance of these data is difficult.
Focus has been put on mapping and annotating each sequence with known genome data. The usual tools for finding specific sequences of the primers, tags, etc. are also available. Final text and graphics statistics completes the programm.
Thus, PostParser is able to process all high-throughput sequencing data in automatic or semiautomatic mode.
This project is still under development and new features or fix are made every month.
Focus has been put on mapping and annotating each sequence with known genome data. The usual tools for finding specific sequences of the primers, tags, etc. are also available. Final text and graphics statistics completes the programm.
Thus, PostParser is able to process all high-throughput sequencing data in automatic or semiautomatic mode.
This project is still under development and new features or fix are made every month.
| To run PostParser on your Unix server, you'll need to use PHP compiler (with the GD library support) and mySQL databases. |
Last updates / new features
01.08.11
Version 2.2b is out! Introducing some fixes on the 2.0 tree. We've also (at least) introduced support for plots. Thanks for your attention!
07.05.11
Some bugs reported from the tracker and related to pie charts have been fixed. Introducing flat pie charts.
|
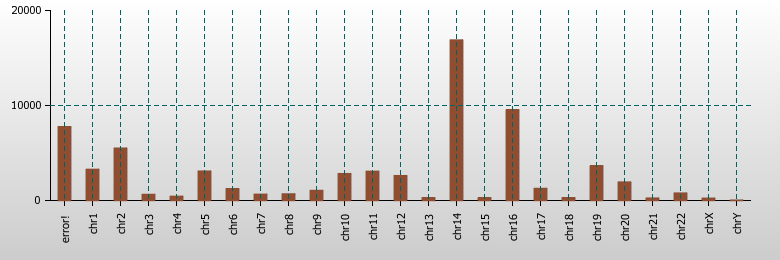 New graphic plots in PostParser |
Documentation
The documentation is trying to explain how to use, adapt and bring to production PostParser software.
Examples are covering basic usage or highlighting specific features, we invite you to try by yourself.
If you can't find any solution to your issues in this documentation, please contact us.
Before main calculating you must understand the Data Origin obtained from sequencing. You'll find all the informations here. To help you dealing with your data transformation we've made special module.
 |
PostParser has been created by Andrew Garazha http://www.pparser.net |